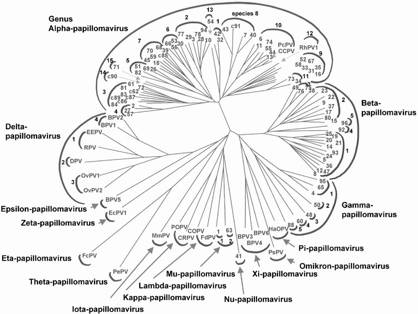
Papillomavirus
classification system
Overview:
|
Taxonomic
level
|
Criteria
|
Number
|
|
Genera
|
Less than
60% nt identity in entire L1 ORF
|
18 genera
(alpha to sigma)
|
|
|
|
2 additional
new genera, yet unnamed (containing TtPV2 and RaPV1 respectively)
|
|
Species
|
Between 60%
and 70% nt identity in L1
|
From 1
species (in most genera) up to 15 species (in genus alpha)
|
|
Genotypes
|
Between 71 and 89 %
nt identity in L1
|
Currently
120 papillomavirus genotypes completely genomically characterized and
available in Genbank (98 human: HPV; 32 non-human)
|
|
Subtypes
|
Between 90
and 98 % identity in L1
|
Very rare
|
|
Variants
|
More than 98
% identity in L1
|
|
Remarks:
-
For a novel
papillomavirus sequence to be classified as a new genotype, the
complete
genome
has to be cloned and the L1 ORF has to differ by more than 10% from the
closest
known PV type.
Genetic
regions suitable for classification:
Current
classification is based on sequence identity in the L1 ORF because:
-
The L1 ORF is
the most conserved gene within the genome, and has been used for
identification
of new PV genotypes over past decades.
-
Almost all
subgenomic PCR amplicons that are generated with the currently
available
consensus or degenerate primer sets are located within the L1 ORF.
Reasons
why
other genomic regions are not used for classification:
-
Some ORFs are
not present in all papillomaviruses: E4, E5, E6 and E7 are lacking in
some PVs.
-
The E1, E2, E4,
E4, E5, E6, E7 and L2 ORFs are all less conserved than L1, making
reliable
alignment of these ORFs difficult for PVs belonging to different
genera, and
only short conserved parts of these ORFs can be reliably aligned.
Reference:
de
Villiers EM,
Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of
papillomaviruses. Virology. 2004 Jun 20;324(1):17-27.