
|
HIV-1 Subtying Tool Methods: The subtyping process consists of four steps:
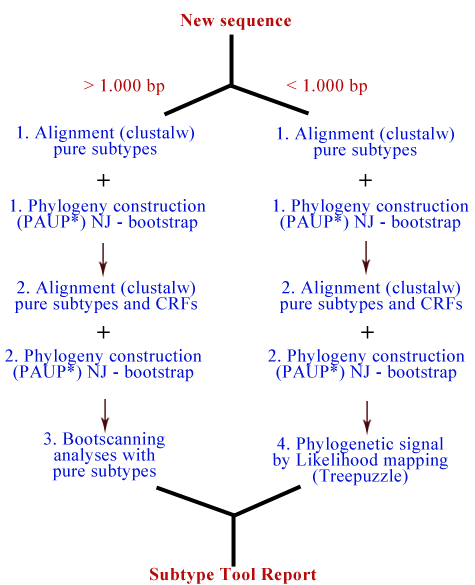 Figure showing the steps used in the HIV-1 subtyping tool. Reference Alignment:
Program software used in the tool:
Developed in cooperation with the Evolutionary Biology Group at University of Oxford, UK., the HIV-1 Pathogenesis and Immunotherapeutics Program at University of Pretoria, South Africa, and the REGA Institute at the Katholieke Universiteit Leuven, Belgium. Funded
by
the Marie Curie Fellowship, Flanders Bilateral Cooperation Program and
the Wellcome
Trust (grant # 061238 ).
For questions, suggestions or
problems please contact: Dr. Tulio
de
Oliveira.
|