
|
REGA HIV-1 Subtying Classification Rules: 1.
Decision Tree for sequences longer than 800 bp:
Back to the
top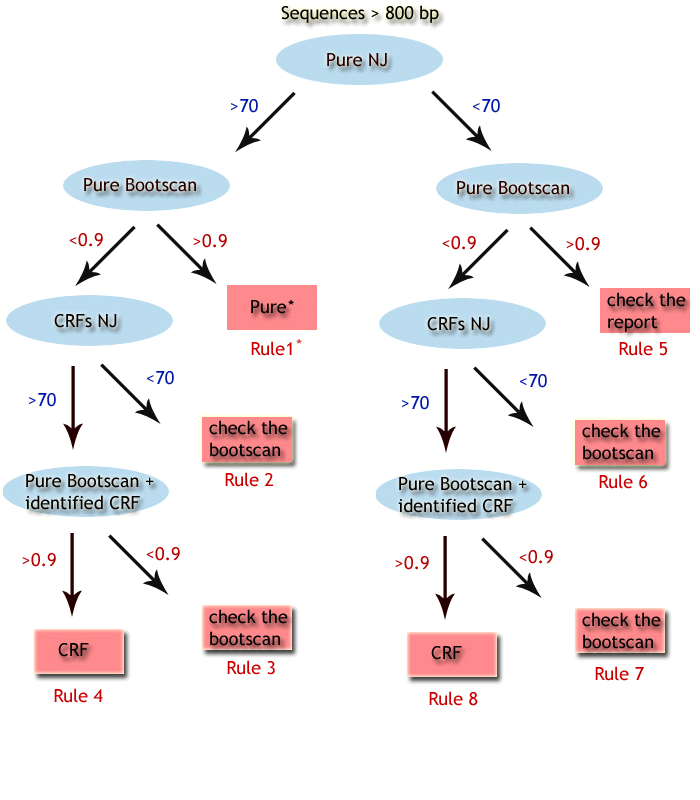 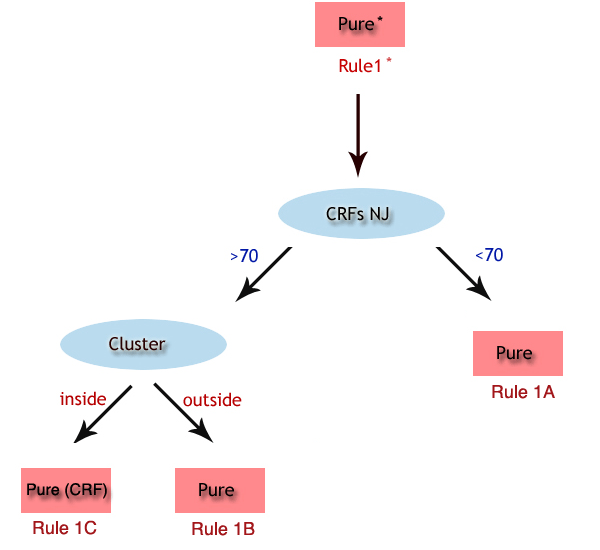 Rule 1A: PURE Subtype assigned based on sequence > 800 bps, clustering with a pure subtype with bootstrap > 70% without recombination in the bootscan, and do not clustering with a CRF with bootstrap >70 %. Rule 1B: PURE Subtype assigned based on sequence > 800 bps, clustering with a pure subtype with bootstrap > 70% without recombination in the bootscan, clustering with a CRF with bootstrap >70 % however not clustering inside the CRF cluster. Rule 1C: PURE (CRF) Subtype assigned based on sequence > 800 bps, clustering with a pure subtype with bootstrap > 70% without recombination in the bootscan, clustering with a CRF with bootstrap >70 % and clustering inside the pure subtype cluster. Rule 2: check the bootscan Subtype unassigned based on sequence > 800bp, clustering with a pure subtype with bootstrap >70 %, with detection of recombination in the pure subtype bootscan, and failure to classify as a CRFs (bootstrap support). Rule 3: check the bootscan Subtype unassigned based on sequence > 800bp, clustering with a pure subtype and CRF with bootstrap >70 %, with detection of recombination in the pure subtype bootscan, and failure to classify as a CRFs (bootscan support). Rule 4: CRF Subtype assigned based on sequence > 800bp, clustering with a CRF with bootstrap > 70%,with detection of recombination in the pure subtype bootscan, and further confirmed as a CRF by bootscan analysis. Rule 5: check the report Subtype unassigned based on sequence > 800bp, do not clustering with a pure subtype with bootstrap >70 %, with no detection of recombination in the pure subtype bootscan. Rule 6: check the bootscan Subtype unassigned based on sequence > 800bp, do not clustering with a pure subtype or CRF with bootstrap >70 %, with detection of recombination. Rule 7: check the bootscan Subtype unassigned based on sequence > 800bp, clustering with a CRF with bootstrap > 70%,with detection of recombination in the pure subtype bootscan, and failure to classify as a CRFs (bootscan support). Rule 8: CRF Subtype assigned based on sequence > 800bp, clustering with a CRF with bootstrap > 70%,with detection of recombination in the pure subtype bootscan, and further confirmed by CRF by bootscan analysis.  Rule 9: check the report Subtype unassigned based on sequence < 800bp, do not clustering with a pure subtype with bootstrap >70 %. Rule 10: check the report Subtype unassigned based on sequence < 800bp, clustering with a pure subtype with bootstrap > 70%, however not clustering inside the pure subtype cluster. Rule 11: Pure Subtype assigned based on sequence < 800bp, clustering with a pure subtype with bootstrap > 70%, and clustering inside the pure subtype cluster
Developed in cooperation with the Evolutionary Biology Group at University of Oxford, UK., the HIV-1 Pathogenesis and Immunotherapeutics Program at University of Pretoria, South Africa, and the REGA Institute at the Katholieke Universiteit Leuven, Belgium. Funded
by
the Marie Curie Fellowship, Flanders Bilateral Cooperation Program and
the Wellcome
Trust (grant # 061238 ).
For questions, suggestions or
problems please contact: Dr. Tulio
de
Oliveira.
|