
ART-AiDE (Antiretroviral Therapy - Aquisition & Display Engine)
Version 1.0, Dec 2006ART-AiDE makes it possible to generate a permanent electronic and graphical record of a patient's antiretroviral treatment (ARV) history, plasma HIV-1 RNA levels, CD4 counts, and (when available) genotypic resistance data.
Submitted data can be reviewed and the underlying XML file can be saved on the Graphical Summary page. The XML file allows you to update a patient's profile and graphical summary without having to re-enter data. The XML schema ART-AiDE uses can be found below:
http://dbpartners.stanford.edu/modules/HIVPatientXML/schema/Complete_Schema.xsd
Save your work frequently because your entries are not stored on our servers after you exit ART-AiDE.
Please report any difficulties using ART-AiDE or desired features to us.
Soo-Yon Rhee
Tommy Liu
Steve Goldman
Jonathan M. Schapiro
Robert W. Shafer
for the Stanford HIV Database
Supported by an unrestricted educational grant from Hoffman-LaRoche.
Table of Contents
- Entry page
- Antiretroviral Therapy (ARVs)
- Antiretroviral Therapy (ARVs) Review
- Plasma HIV-1 RNA & CD4+ Lymphocytes (RNA/CD4)
- Graphical Summary
- Sequences
- HIV db Interpretation
- Save Data To Computer
Entry page
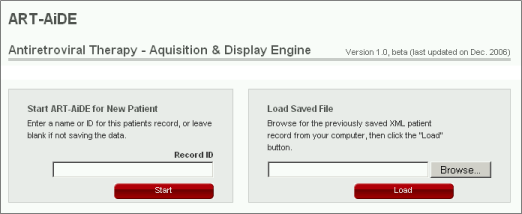
ART-AiDE is begun by entering data on a new patient or by uploading a file previously created by the ART-AiDE program. For example, when you save your work, it will be saved in as an XML file that you can store on your computer and update without having to re-enter data.
To start ART-AiDE for a new patient, press the button "Start" in the left grey box. The 'Record ID' is optional. However, if you plan to print the graphical output or to save the XML for future analyses, then it will be useful to enter a unique 'Record ID'.
To load previously entered data, press the button "Browse", choose the file on your computer and press the button "Load" in the right grey box. On pressing "Load", ART-AiDE will first check if the format of the file you upload is in the XML schema (http://dbPartners.stanford.edu/DDCRP_UI/schema/Complete_Schema.xsd). If the file is in the XML schema, the data in the file will be processed and loaded to ART-AiDE. The ART-AiDE schema is an open source schema that is being widely adopted for representing patient antiretroviral treatments, virus levels, CD4 counts, and drug resistance data. Although the ART-AiDE schema will be updated over time, a version control system guarantees that previously created files will always be readable and updatable to the most recent XML version.
Antiretroviral Therapy (ARVs)
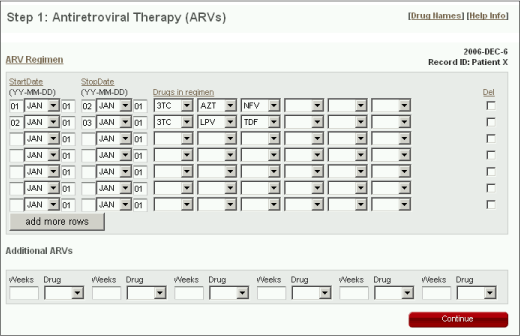
ARV Regimen
The Antiretroviral Therapy (ARV) form is the first form encountered when entering new patient data.
- Each row is designed to contain an ARV regimen or set of simultaneously administered drugs.
- The start and stop dates for each regimen must be entered. The 'StartDate' and 'StopDate' each consist of year (2 digits), month (drop-down menu), and day (2 digits).
- To enter more than six drugs in a single regimen, continue the regimen on a new line that contains the same 'StartDate' and 'StopDate'.
- The regimens do not need to be entered in order but they will be saved in temporal order after leaving this page.
- The 'Del' check box is used for editing purposes to delete a regimen that has already been entered.
- ARVs are selected from a drop-down list that contains all FDA-licensed drugs, "None", and 5 categories to represent varying degrees of uncertainty: "Unknown" indicates that one or more drugs may have been received but that nothing is known about it. "RTI" indicates that one or more RT inhibitors were received but the specific RTIs are not known. "NRTI" indicates that one or more NRTIs were received but the specific RTIs. "NNRTI" indicates that one or more NNRTIs were received but the specific RTIs. "PI" indicates that a PI was received by that the specific PI is not known.
- There should be no gaps between regimens. If treatment was not used, select "None" from the drop-down menu. If an ARV regimen is unknown, select "Unknown" from the drop-down menu.
- ARVs can also be selected by entering the first letter of the drug after placing the cursor in the drop-down box. The tab can be used to move between drugs and regimens.
Additional ARVs
Use the "Additional ARV" section to indicates drugs known to be received but for which the simultaneously administered drugs, the dates of administration, and the duration of administration are not known. Therefore, it is only necessary to enter the drug name. Entering the duration of therapy with the drug is optional.
Navigation
After the ARV treatment history has been entered or updated, press "Continue" to go to a page where the treatment history can be reviewed graphically.
This form will be updated to allow users to enter investigational ARVs and to simplify the entering of fixed-dose combinations and boosted PIs.
Antiretroviral Therapy (ARVs) Review
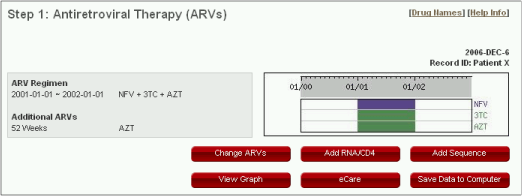
Navigation
- "Change ARVs" to go back to the ARV page for further updates
- "Add RNA CD4" to add plasma HIV-1 RNA level or CD4+ lymphocytes counts (see the section "Plasma HIV-1 RNA & CD4+ Lymphocytes")
- "Add Sequence" to add genotype testing results (see the section "Sequences")
- "View Graph" to generate graphical summary of the ARVs, if available, plasma HIV-1 RNA levels, CD4 counts and drug resistance mutations (see the section "Graphical Summary").
- "eCare" to browse the patterns of drug resistance mutations in the Stanford Drug Resistance Database obtained from persons receiving the same ARVs entered (see the section "eCARE").
- "Save Data To Computer" to save the data entered in a XML file on your computer (see the section "Save Data To Computer").
Plasma HIV-1 RNA & CD4+ Lymphocytes (RNA/CD4)
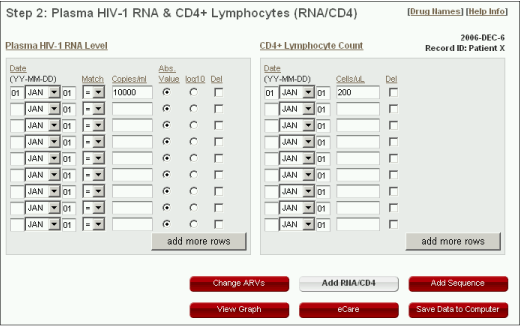
Plasma HIV-1 RNA Level
- The date for each RNA measurement must be entered. The dates consist of three parts: year, month, and day. The year should be entered using two digits. The month is selected from a drop-down list. The day must be typed in but can be left at '01' if it is not known.
- "Match" is used to indicate exact RNA levels ("=") and levels below ("<") the lower limit of detection or above (">") the upper limit of quantification.
- The number of RNA copies/ml can be entered either as an absolute value or as a log (base 10) value. The radio buttons ("Abs. Value" and "log10") can be used to indicate whether the value is an absolute or a log based.
CD4+ Lymphocyte count
- The date for each RNA measurement must be entered. The dates consist of three parts: year, month, and day. The year should be entered using two digits. The month is selected from a drop-down list. The day must be typed in but can be left at '01' if it is not known.
- The absolute CD4 count (cells/µL) should be entered.
Navigation
- "Change ARVs" to go back to the ARV page for further updates
- "Add Sequence" to add genotype testing results (see the section "Sequences")
- "View Graph" to generate graphical summary of the ARVs, plasma HIV-1 RNA levels and CD4 counts that have been updated or entered and, if available, drug resistance mutations (see the section "Graphical Summary").
- "eCare" to browse the patterns of drug resistance mutations in the Stanford Drug Resistance Database obtained from persons receiving the same ARVs (see the section "eCARE").
- "Save Data to Computer" to save the data entered in a XML file on your computer (see the section "Save Data To Computer").
We are developing a batch upload system for RNA levels and CD4 counts in a tab-delimited text file.
Graphical Summary
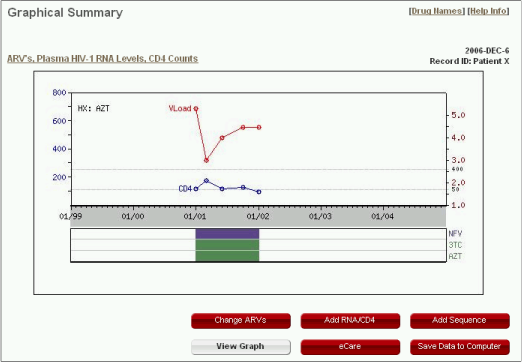
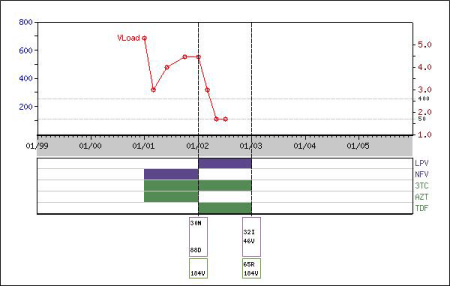
The graphical output illustrates the temporal relationship between the ARVs, plasma HIV-1 RNA levels and CD4 counts. The figure is updated as changes are made using the forms.
- Protease inhibitors are purple, nucleoside RT inhibitors are green, and non-nucleoside RT inhibitors are cyan. "HX" indicates ARVs received but for which the duration or the simultaneously administered ARVs are not known.
- The log RNA levels (red dots) are indicated along the right-sided Y-axis. Inverted triangles indicate levels below the level of detection.
- The CD4 counts (blue dots) are indicated using the left-sided Y-axis.
When genotype testing results was entered using the form "Add Sequence" (see the section "Sequence"), the test results will be shown as a list of major drug resistance mutations as below: a list of protease drug resistance mutations inside a purple rectangle; a list of RT drug resistance mutations inside a green rectangle.
Sequences
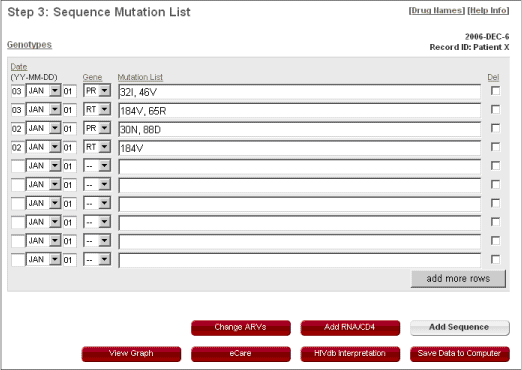
- Each row is designed to contain the mutation list belonging to a gene sequence (PR or RT) specified in the "Gene" drop-down list.
- The date for each gene sequence must be entered. The dates consist of three parts: year, month, and day. The year should be entered using two digits. The month is selected from a drop-down list. The day must be typed in but can be left at '01' if it is not known.
- To use the "Mutation List" text box, type each mutation upper case separated by one or more spaces (the consensus wildtype amino acids and separating commas are optional). If there is a mixture of more than one amino acid at a position, write both amino acids (an intervening slash is optional). Use lowercase "i" to indicate an insertion and lowercase "d" to indicate a deletion.
- If you want to delete one or more gene sequences, check the "Del" selection box/es for the appropriate gene sequence/s and date/s you want to delete.
- Click on "View Graph" to see the sequence updates.
- Click on "HIVdb Interpretation" to apply the original (non-eCARE) Stanford HIVdb algorithm on an individual sequence or set of sequences.
HIV db Interpretation
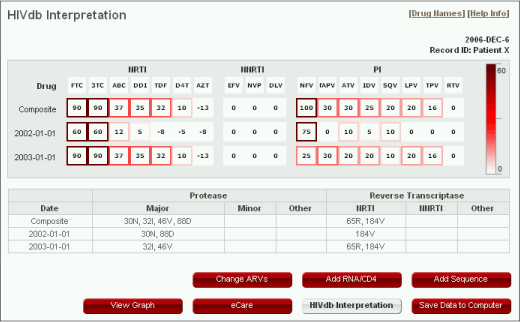 >
>Each row shows the inferred levels of resistance to individual FDA-approved ARVs according to as explained in the program release notes (http://hivdb.stanford.edu/pages/algs/HIVdb.html). However, unlike the main program, it calculates scores for individual sequences as well as scores for multiple sequences obtained by pooling mutations observed at all time points. Note: This program does not consider ARV treatment history.
Save Data To Computer
The ARV treatment, RNA levels, CD4 counts and genotype testing results that have been entered can be saved on your computer as a XML file. On pressing the button "Save Data to Computer", the data will be downloaded to your browser. Select "Save As" under the File menu on the browser to save the data on your computer as a file. Specify the name of file with a ".xml" extension. Once the data saved in a file, you can resume the ART-AiDE by uploading the file on the "Entry page". In this way, you can update a patient's profile and graphical summary without having to re-enter data.